Fundamental biological mechanisms such as development, maintenance of homeostasis, cell differentiation, and adaptation in various environmental contexts (normal or pathological) are controlled by the fine regulation of genetic networks over time and space at the single-cell level.
For technical reasons, biologists have long been limited to observe the average transcriptome of groups of cells within a tissue or organ. Tissues are, however, made up of individual cells at various stages with a wide variety of shapes and functions. Recent developments in single-cell sequencing have revealed an unexpected heterogeneity in the transcriptome of neighboring cells, thereby also opening many new questions (transcriptome homeostasis, gene expression robustness, transcriptional noise filtering, spatio-temporal averaging of cell identities, etc.).
It is now essential to systematically characterize the spatial distribution of this heterogeneity and its dynamics over time in situ. The in vivo characterization of each cell's transcriptome in its biological environment, called “spatial transcriptomics”, coined “the 2018 breakthrough of the year” according to Science, is about to revolutionize our understanding of life.
Spatial-Cell-ID aims to identify and characterize the transcriptome of each cell and its dynamics in animal and plant organisms, whether adult or in development, healthy or sick. For this purpose, we are creating a national spatial transcriptomics equipment.
This equipment will integrate the latest developments in spatial transcriptomics, some of which are not commercialized yet, for small to large numbers of transcripts, in intact or sorted cells, in whole organisms or tissue sections, combined with data analysis. Such a wide spectrum of cutting-edge approaches working in synergy on the same site is unprecedented in Europe.
Given the amount of data produced, the integration of a high-performance computational infrastructure is essential. The breakthrough behind Spatial-Cell-ID ID is to determine the transcriptome of each cell in tissues, either through data acquisition in situ at high resolution, or through algorithmic repositioning of the cell into the tissue.
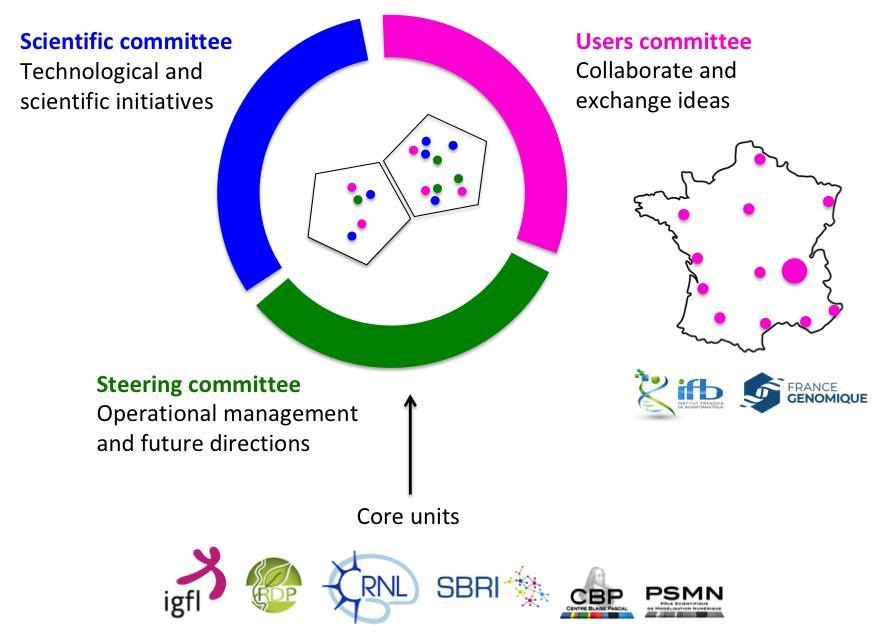
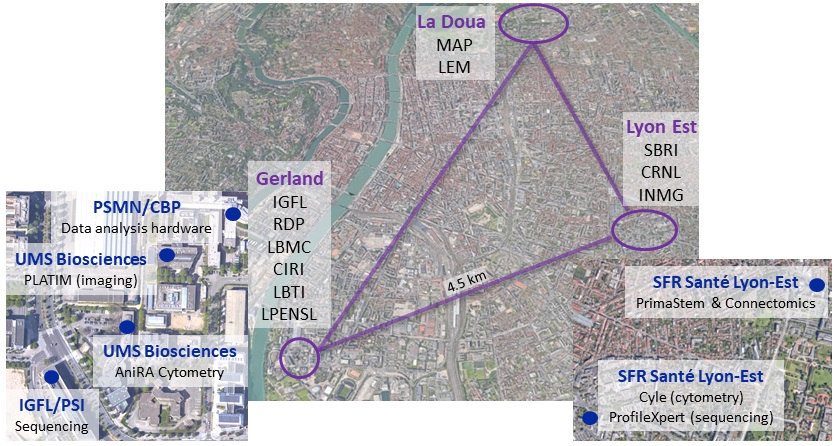
Spatial-Cell-ID is the first facility in France to provide integrated single-cell spatio-temporal analysis in strong relation with the national network of genomics, imaging, and bioinformatics infrastructures “France Génomique”, “France BioImaging”, and IFB cloud (“Institut Français de Bioinformatique”).
Scientific coordinators
- Yad Ghavi-Helm (IGFL - Genomics, UMR5242)
- Jonathan Enriquez (IGFL - Neurodevelopment, UMR5242)
- Marie Monniaux (RDP – Plant development, UMR5667)
Project manager
Romain Guyot (IGFL – Genomics, UMR5242)
Administrative and financial manager
Fabienne Rogowsky (IGFL – UMR5242)